Detección de serotipos y genes de resistencia antimicrobiana de Streptococcus pneumoniae en muestras clínicas de pacientes con enfermedad invasiva en Paraguay
Contenido principal del artículo
Resumen
Introducción: Streptococcus pneumoniae es el agente etiológico responsable de enfermedades invasivas graves como neumonías bacterianas adquiridas en la comunidad, sepsis y meningitis. Sigue siendo una de las causas más importantes de morbilidad y mortalidad en niños y adultos alrededor del mundo. Los datos deserotipos y perfil de resistencia antimicrobiana proporcionaran información relevante para fundamentar las directrices de tratamiento y las políticas de vacunación.
Objetivos: detectar serotipos y genes de resistencia antimicrobiana en muestras clínicas de niños y adultos con enfermedad invasiva causada por Streptococcuspneumoniae en Paraguay en el año 2023.
Metodología: estudio descriptivo retrospectivo de corte transversal. Se estudiaron todas las muestras clínicas (n = 145) de pacientes con enfermedad invasiva por Streptococcus pneumoniaeque no fueron asiladas en cultivos bacteriológicos provenientes de los centros centinela y colaboradores en el marco de la vigilancia de meningitis y neumonías de Paraguay durante el año 2023.
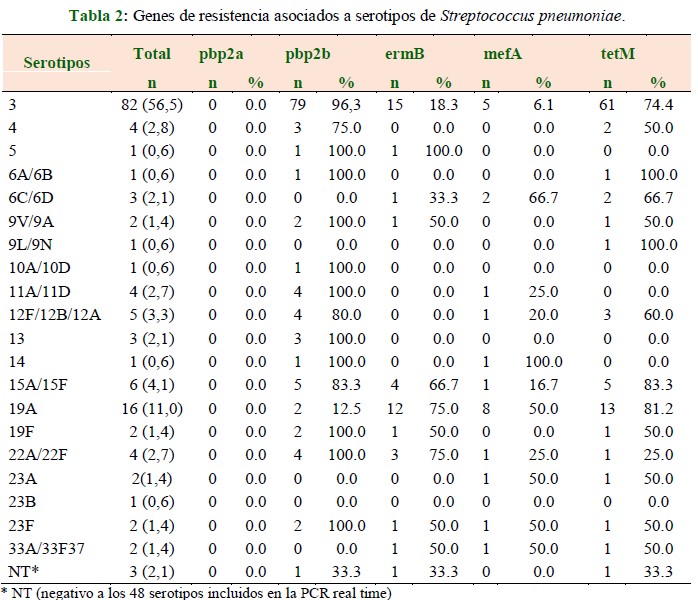
Resultados: se identificaron 20 serotipos, siendo los más frecuentes el serotipo 3 en 56,5 %, el 19A en 11,0 % y el 15A/15F en 4,1 %. En el serotipo 3 se detectó el gen pbp2b en 96,3 %, ermB 18,3 %, mefA en 6,1 % y tetMen 74,4 %, en el serotipo 19A el gen pbp2b en 12,5 %, ermB en 75,0 %, mefA en 50,0 % y tetMen 81,2 %, el serotipo 15A/15F el gen pbp2b en 83,3 %, ermBen 66,7 %, mefA en 16,7 % y tetM en 83,3 %.
Conclusión: se detectó un alto porcentaje de pbp2b lo que indica una alta susceptibilidad a la penicilina, sin embargo, también se observó alta resistencia a tetraciclina relacionada con el serotipo 3 y resistencia a macrólidos relacionada a los serotipos 19A y 15A/15F. Técnicas moleculares aplicadas directamente al material clínico tienen el potencial de detectar el organismo infectante y determinar su susceptibilidad a los antimicrobianos, facilitando así la terapia oportuna e intervención con antibióticos adecuados.
Detalles del artículo

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Usted es libre de:
- Compartir: copiar y redistribuir el material en cualquier medio o formato para cualquier propósito, incluso comercialmente.
- Adaptar: remezclar, transformar y construir a partir del material para cualquier propósito, incluso comercialmente.
- La licenciante no puede revocar estas libertades en tanto usted siga los términos de la licencia
Bajo los siguientes términos:
- Atribución: Usted debe dar crédito de manera adecuada, brindar un enlace a la licencia, e indicar si se han realizado cambios. Puede hacerlo en cualquier forma razonable, pero no de forma tal que sugiera que usted o su uso tienen el apoyo de la licenciante.
- Compartir igual: — Si remezcla, transforma o crea a partir del material, debe distribuir su contribución bajo la misma licencia del original.
- No hay restricciones adicionales — No puede aplicar términos legales ni medidas tecnológicas que restrinjan legalmente a otras a hacer cualquier uso permitido por la licencia.
Citas
Cedrone F, Montagna V, Del Duca L, Camplone L, Mazzocca R, Carfagnini F, et al. The burden of Streptococcus pneumoniae-related admissions and in-hospital mortality: a retrospective observational study between the years 2015 and 2022 from a Southern Italian Province. Vaccines(Basel). 2023;11(8):1324. doi: 10.3390/vaccines11081324.
Thadchanamoorthy V, Dayasiri K. Review on pneumococcal infection in children. Cureus. 2021;13(5): e14913. doi: 10.7759/cureus.14913.
Martinez-Vega R, Jauneikaite E, Thoon KC, Chua HY, HuishiChua A, Khong WX, et al. Risk factor profile sand clinical outcomes for children and adults with pneumococcal infections in Singapore: a need to expand vaccination policy? PLoS ONE. 2019;14(10): e0220951. doi: 10.1371/journal.pone.0220951.
Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Global Health. 2018;6(7): e744–57. doi: 10.1016/S2214-109X(18)30247-X.
GBD 2015 LRI Collaborators. Estimates of the global, regional, and national morbidity, mortality, and etiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17(11): 1133–61. doi: 10.1016/S1473-3099(17)30396-1.
Ganaie F, Saad JS, McGee L, van Tonder AJ, Bentley SD, Lo SW, et al. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral streptococcus. ASM Journals. mBio. 2020;11(3):e00937-20. doi: 10.1128/mBio.00937-20.
Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16(6):355–367. doi: 10.1038/s41579-018-0001-8.
Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Disponible en: https://www.cdc.gov/antimicrobial-resistance/media/pdfs/2019-ar-threatsreport-508.pdf
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet. 2022;399(10325):629–55. doi: 10.1016/S0140-6736(21)02724-0.
Li L, Ma J, Yu Z, Li M, Zhang W, Sun H. Epidemiological characteristics and antibiotic resistance mechanisms of Streptococcus pneumoniae: an updated review. Microbiol Res. 2023;266:127221. doi: 10.1016/j.micres.2022.127221.
Ktari S, Ben Ayed N, Ben Rbeh I, Garbi N, Maalej S, Mnif B, et al. Antibiotic resistance pattern, capsular types, and molecular characterization of invasive isolates of Streptococcus pneumoniae in the south of Tunisia from 2012 to 2018. BMC Microbiol. 2023;23(1):36. doi: 10.1186/s12866-023-02784-2.
León ME, Samudio M, Kawabata A, Nagai M, Rojas L, Zárate N, et al. Serotypes and antimicrobial resistance of Streptococcus pneumoniae in children under 5 years of age pre- and post-pneumococcal conjugate vaccine introduction in Paraguay. J Med Microbiol. 2023;72(6):001700. doi: 10.1099/jmm.0.001700.
Souza MB, Cergole-Novella MC, Molinari DA, Colpas DR, Carmo AMdS, Daros VdSMG, et al. Multiplex real-time PCR using SYBR Green: unspecific intercalating dye to detect antimicrobial resistance genes of Streptococcus pneumoniae in cerebrospinal fluid. PLoS ONE. 2022;17(6): e0269895. doi: 10.1371/journal.pone.0269895.
Ouattara M, Whaley MJ, Jenkins LT, Schwartz SB, Traoré RO, Diarra S, et al. Triplex real-time PCR assay for the detection of Streptococcus pneumoniae, Neisseria meningitidis and Haemophilus influenzae directly from clinical specimens without extraction of DNA. Diag Microbiol Infect Dis. 2019;93(3):188-190. doi: 10.1016/j.diagmicrobio.2018.10.008.
Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44(1):124-31. doi: 10.1128/JCM.44.1.124-131.2006.
Velusamy S, Tran T, Mongkolrattanothai T, Walker H, McGee L, Beall B. Expanded sequential quadriplex real-time polymerase chain reaction (PCR) for identifying pneumococcal serotypes, penicillin susceptibility, and resistance markers. Diagn Microbiol Infect Dis. 2020;97(2):115037. doi: 10.1016/j.diagmicrobio.2020.115037.
Pizzutti K, Perez VP, Barbiero C, d'Azevedo PA, Fischer GB, Dias C. Identifying pneumococci in parapneumonic pleural effusion: is there a role for culture-independent methods? Pediatric Pulmonol. 2020;55(2): 484–489. doi: 10.1002/ppul.24568.
Silva-Costa C, Gomes-Silva J, Pinho MD, Friães A, Ramirez M, Melo-Cristino J. Continued vaccine breakthrough cases of serotype 3 complicated pneumonia in vaccinated children, Portugal (2016–2019). Microbiol Spectr. 2022;10(4):e01077-22. doi: 10.1128/spectrum.01077-22.
Strachan R, Homaira N, Beggs S, Bhuiyan MU, Gilbert GL, Lambert SB, et al. Assessing the impact of the 13 valent pneumococcal vaccine on childhood empyema in Australia. Thorax. 2021; 76(5):487–493. doi: 10.1136/thoraxjnl-2020-216032.
Goettler D, Streng A, Kemmling D, Schoen C, von Kries R, Rose MA, et al. Increase in Streptococcus pneumoniae serotype 3 associated parapneumonic pleural effusion/empyema after the introduction of PCV13 in Germany. Vaccine. 2020;38(3):570–577. doi: 10.1016/j.vaccine.2019.10.056.
Cillóniz C, Garcia-Vidal C, Ceccato A, Torres A. Antimicrobial Resistance Among Streptococcus pneumoniae. Antimicrobial Resistance in the 21st Century. 2018;7:13–38. doi: 10.1007/978-3-319-78538-7_2.
Almeida SCG, Lemos APS, Bierrenbach AL, Moraes JC, Brandileone MCC. Serotype Distribution and Antimicrobial Susceptibility Pattern of Streptococcus pneumoniae in COVID-19 Pandemic Era in Brazil. Microorganisms. 2024; 17;12(2):401. doi: 10.3390/microorganisms12020401.
Camacho-Moreno G, Leal AL, Patiño-Niño J, Vasquez-Hoyos P, Gutiérrez I, Beltrán S, et al. Serotype distribution, clinical characteristics, and antimicrobial resistance of pediatric invasive pneumococcal disease in Colombia during PCV10 mass vaccination (2017–2022). Front Med (Lausanne). 11:1380125. doi: 10.3389/fmed.2024.1380125.
Izquierdo C, Ciruela P, Hernández S, García-García JJ, Esteva C, Moraga-Llop F, et al. Pneumococcal serotypes in children, clinical presentation and antimicrobial susceptibility in the PCV13 era. Epidemiol Infect. 2020;148:e279. doi:10.1017/S0950268820002708.
Kwun MJ, Ion AV, Cheng HC, D`Aeth JC, Dougan S, Oggiioni MR, et al. Post-vaccine epidemiology of serotype 3 pneumococci identifies transformation inhibition through prophagedriven alteration of a non-coding RNA. Genome Med. 2022;14(1):144. doi: 10.1186/s13073-022-01147-2.
Savulescu C, Krizova P, Valentiner-Branth P, Ladhani S, Rinta-Kokko H, Levy C, et al. Effectiveness of 10 and 13-valent pneumococcal conjugate vaccines against invasive pneumococcal disease in European children: SpIDnet observational multicentre study. Vaccine. 2022; 40(29):3963-3974. doi: 10.1016/j.vaccine.2022.05.011.
Pérez-Abeledo M, Zaragoza G, Ramos B, Sanz JC. High frequency of Streptococcus pneumoniae serotype 3 in negative pleural fluid cultures from paediatric samples obtained in the Madrid region from 2018 to 2022, detected by direct identification using PCR-reverse-hybridization strip-based assay. Enferm Infecc Microbiol Clin (EIMC). 2023;41(7): 447-448. doi: 10.1016/j.eimce.2023.02.005.
Zahari NIN, EngkuAbd Rahman ENS, Irekeola AA, Ahmed N, Rabaan AA, Alotaibi J, et al. A Review of the resistance mechanisms for β-lactams, macrolides and fluoroquinolones among Streptococcus pneumoniae. Medicina (Kaunas). 2023;31;59(11):1927. doi: 10.3390/medicina59111927.
Hasanuzzaman M, Malaker R, Islam M, Baqui AH, Darmstadt GL, Whitney CG, et al. Detection of macrolide resistance genes in culture-negative specimens from Bangladeshi children with invasive pneumococcal diseases. J Glob Antimicrob Resist. 2017;8:131–134. doi:10.1016/j.jgar.2016.11.009.
Zhou X, Liu J, Zhang Z, Cui B, Wang Y, Zhang Y, et al. Characterization of Streptococcus pneumoniae macrolide resistance and its mechanism in northeast china over a 20-year period. Microbiol Spectr. 2022;10(5):e00546-22. doi: 10.1128/spectrum.00546-22.
Gonzales BE, Mercado EH, Pinedo-Bardales M, Hinostroza N, Campos F, Chaparro E, et al. Increase of macrolide-resistance in Streptococcus pneumoniae strains after the introduction of the 13-valent pneumococcal conjugate vaccine in Lima, Peru. Front Cell Infect Microbiol. 2022;12:866186. doi: 10.3389/fcimb.2022.866186.
Beheshti M, Jabalameli F, Feizabadi MM, Bonakdar Hahsemi F, Beigverdi R, Emaneini M. Molecular characterization, antibiotic resistance pattern and capsular types of invasive Streptococcus pneumoniae isolated from clinical samples in Tehran, Iran. BMC Microbiol. 2020;20(1):167. doi: 10.1186/s12866-020-01855-y.
Gagetti P, Lo SW, Hawkins PA, Gladstone RA, Regueira M, Faccone D, et al. Population genetic structure, serotype distribution and antibiotic resistance of Streptococcus pneumoniae causing invasive disease in children in Argentina. Microb Genom. 2021;7(9):000636. doi: 10.1099/mgen.0.000636.
Gagetti P, Faccone D, Reijtma V, Fossati S, Rodriguez M, Veliz O, et al. Characterization of Streptococcus pneumoniae invasive serotype 19A isolates from Argentina (1993–2014). Vaccine. 2017;35(35):4548-4553. doi: 10.1016/j.vaccine.2017.07.030.
Nakao T, Kosai K, Akamatsu N, Ota K, Mitsumoto-Kaseida F, Hasegawa H, Izumikawa K, et al. Molecular and phenotypic characterization of Streptococcus pneumoniae isolates in a Japanese tertiary care hospital. Front Cell Infect Microbiol. 2024;14:1391879. doi: 10.3389/fcimb.2024.1391879.